Explore the world through data.

I am a statistician experienced with genomics (population, neuroimaging, and single-cell genetics), biobank and phenotypic data, sports analytics, and administrative and census databases. I am currently a postdoctoral researcher at the Data Science Institute at Brown University, where I hold a Postdoctoral Fellowship from the Natural Sciences and Engineering Research Council of Canada (NSERC).
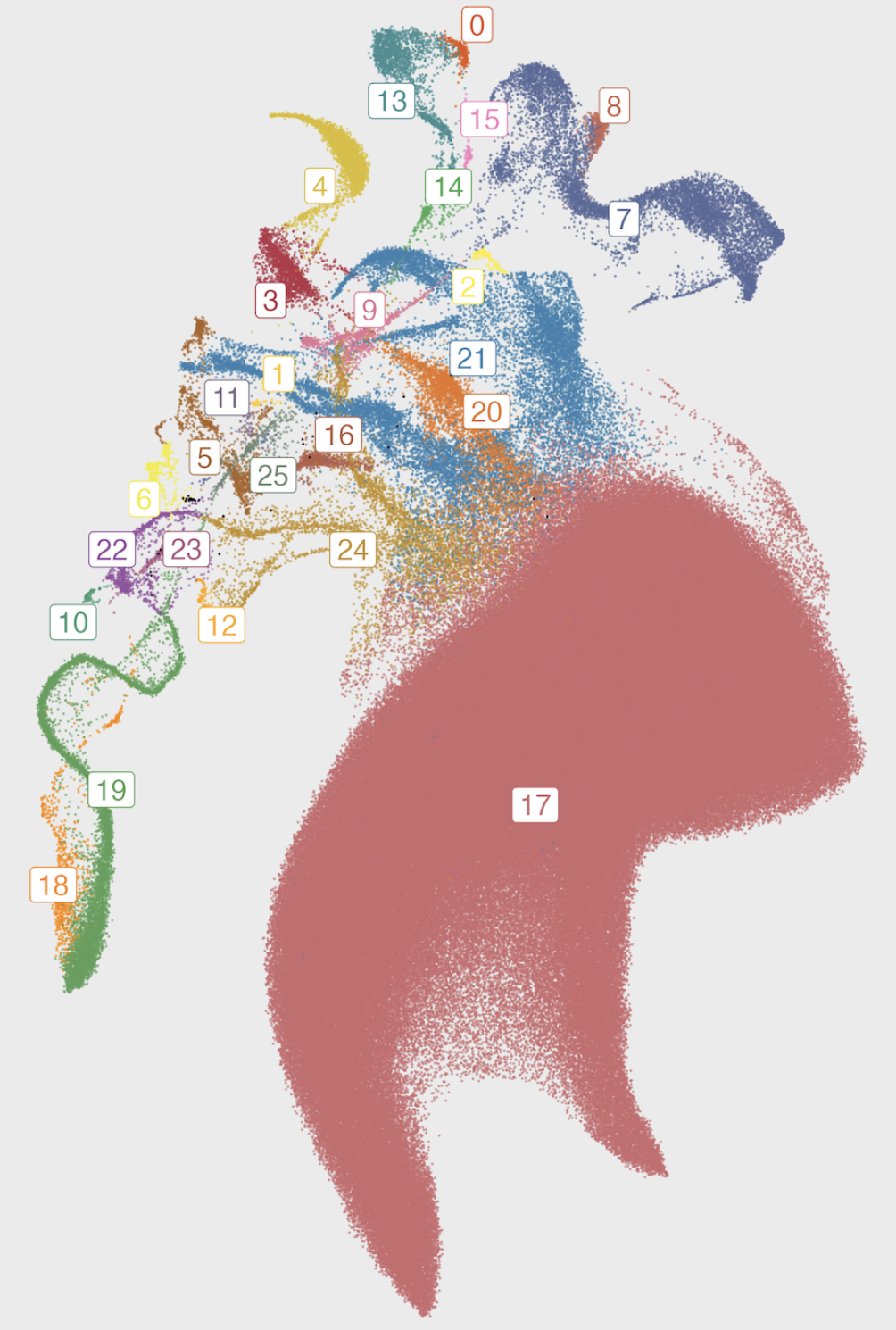
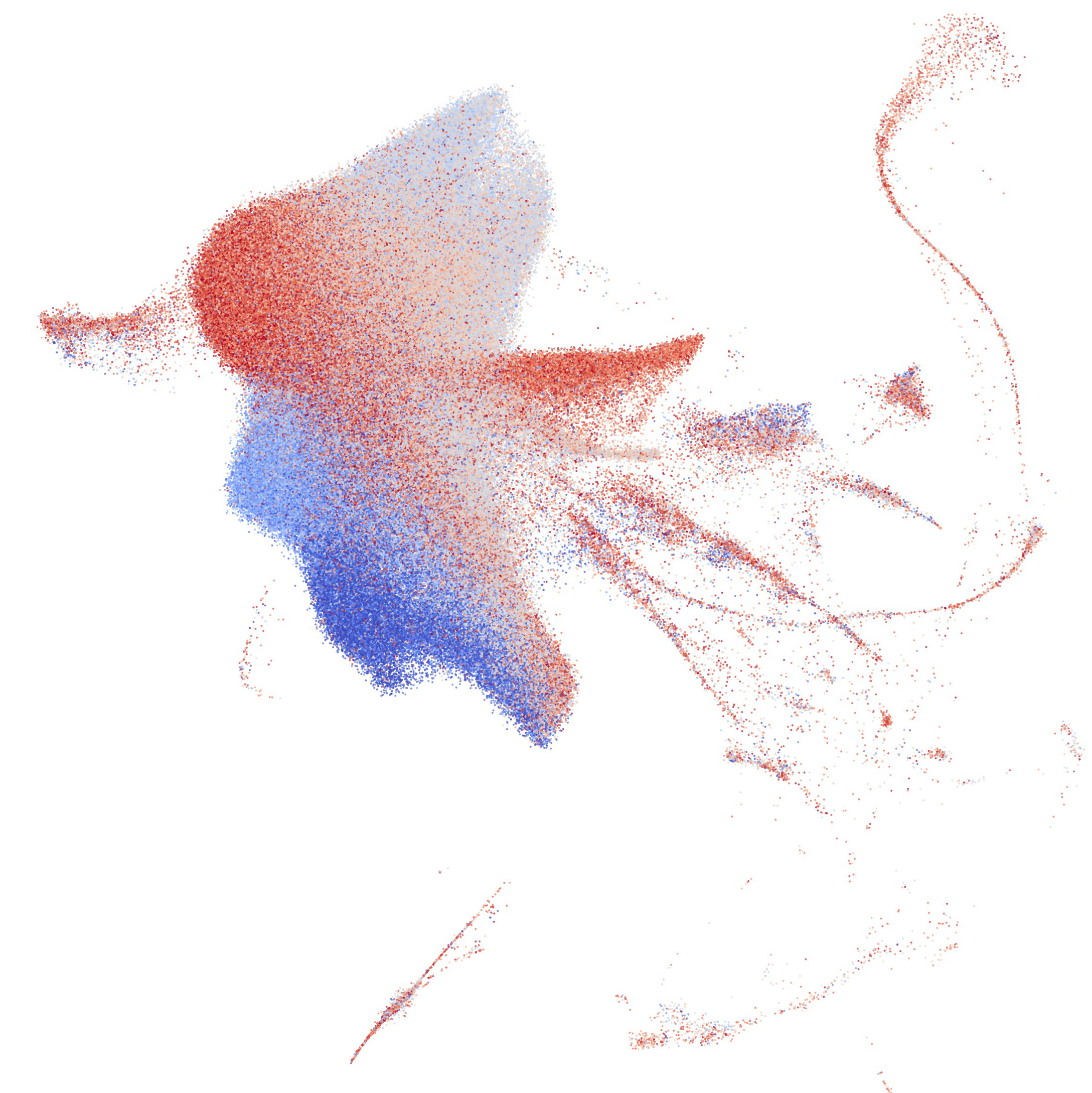
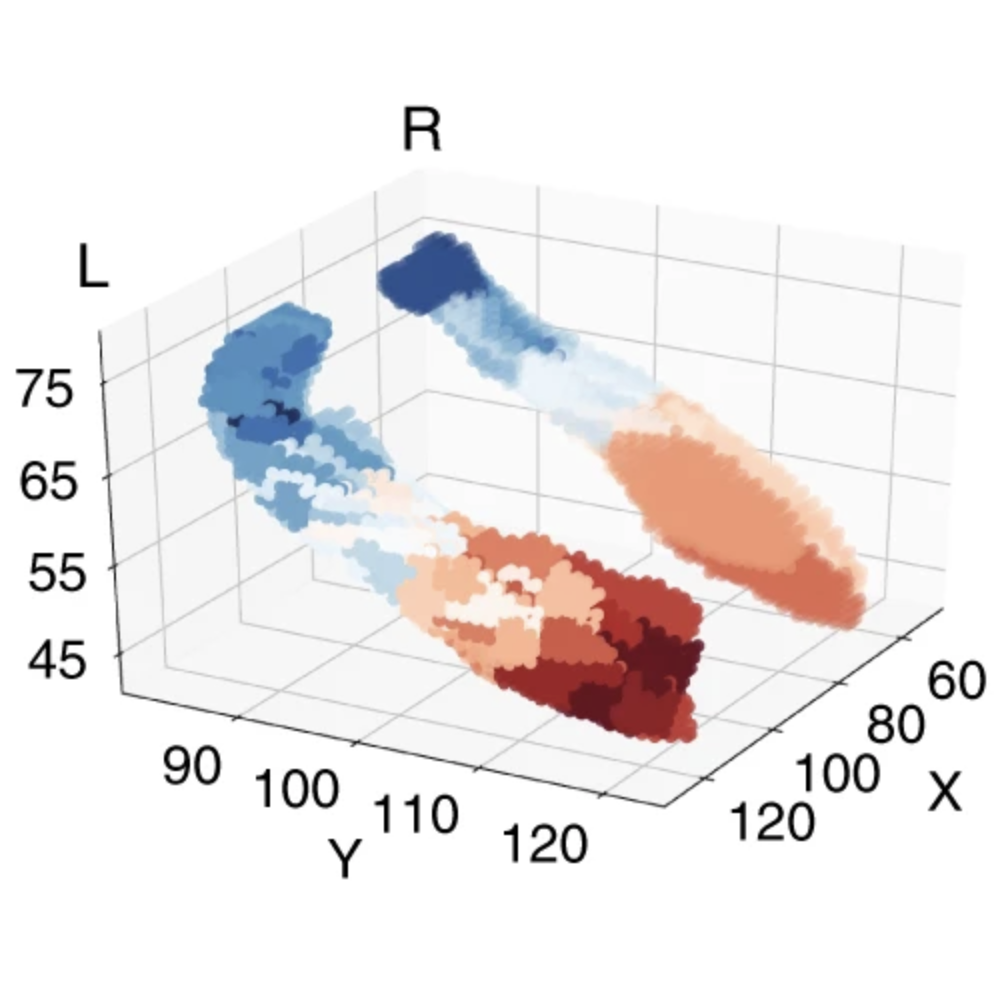
My research has included statistical machine learning methods in dimension reduction, visualization, and clustering of genomic data. I also have expertise in genome-wide association analysis, polygenic scoring, and statistical genetics tools (ADMIXTURE, PLINK, etc) and high-performance and cloud computing environments. I have studied biobanks including the 1000 Genomes Project, CARTaGENE, the UK biobank, and AllofUs.
My primary research interest is developing and applying methods for high-dimensional data. I like to study the distributions of phenotypes and environmental variables in large and diverse biobanks through the lens of genetics (e.g. through population structure) and seeing how the distributions differ. I am also interested in studying the social impacts of human genetics research, such as how decades of genomics and consumer genetic testing shape how we think of and define populations.
I enjoy statistics and machine learning for personal projects, such as traffic safety. Among these are projects using computer vision to track traffic and collecting news stories of pedestrian collisions for further statistical analyses.
My main tools and skills:I worked as a methodologist on a variety of surveys and technical projects, including the International Travel Survey, Canadian Income Survey, and developing the Longitudinal Immigration Database. Among my responsibilities were:
I worked in several roles on an ad-hoc basis. These included:
The Entangled Biobank: On the Topology of High-Dimensional Human Genetic Data
Data Mining the Play-by-Play: Assessing and Applying NHL Performance Metrics Using Statistical Methods